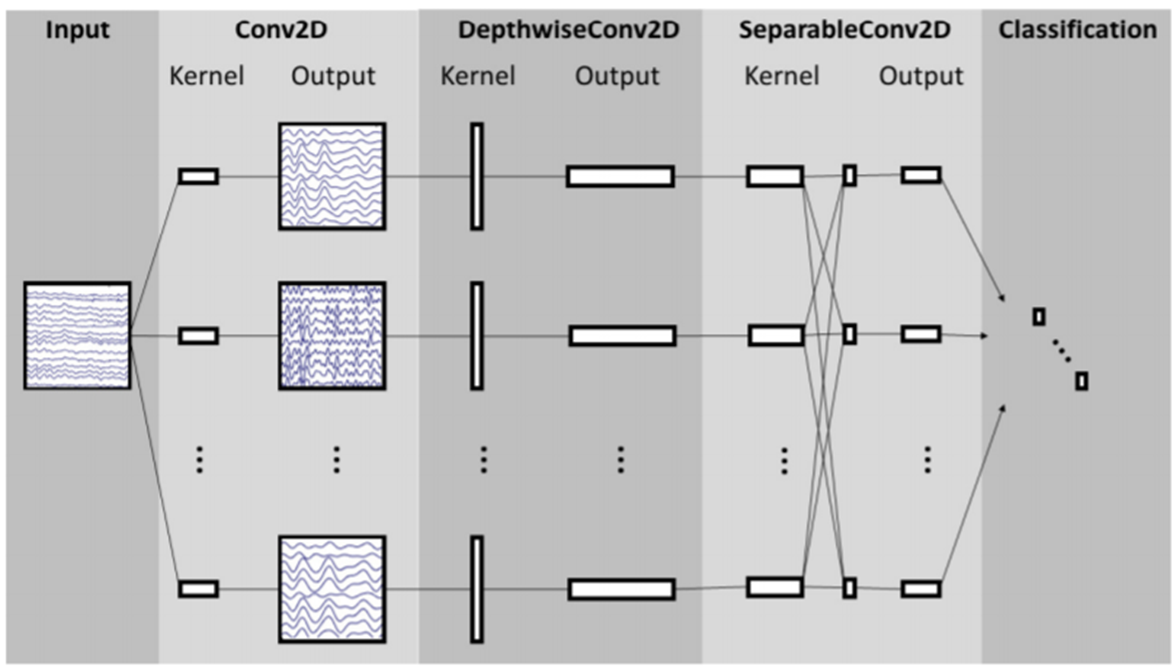
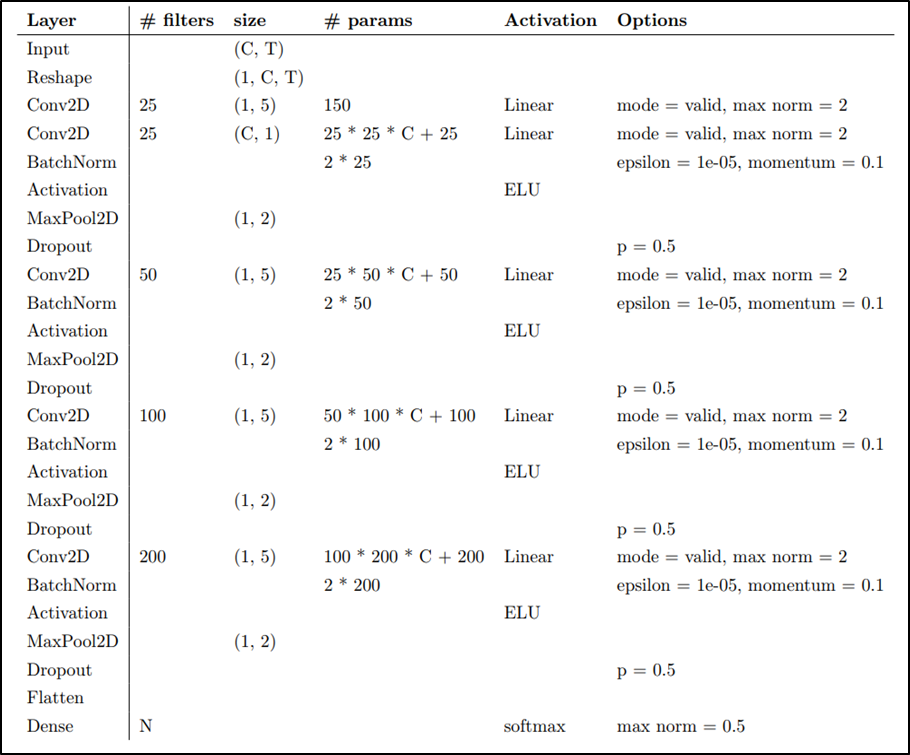
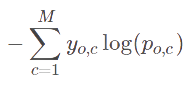The task is to classify BCI competition datasets(EEG signals) by using EEGNet and DeepConvNet with different activation functions. I have built EEGNet and DeepConvNet by using pytorch.
You can get some detailed introduction and experimental results in this link.
Operating System: Windows 10
CPU: Intel(R) Core(TM) i7-6700 CPU @ 3.40GHz
GPU: NVIDIA GeForce GTX TITAN X
In this work, you can use the following two option to build the environment.
$ conda env create -f environment.yml$ conda create --name Summer python=3.8 -y
$ conda activate Summer
$ conda install pytorch==1.7.1 torchvision==0.8.2 torchaudio==0.7.2 cudatoolkit=10.2 -c pytorch
$ conda install numpy
$ conda install matplotlib -y
$ conda install pandas -y
$ pip install torchsummary
The model architecture that combines with different activation function was in the ALL_model.py file.
In this project, the training and testing data were provided by BCI Competition III – IIIb.
Data: [Batch Size, 1, 2, 750]
Label: [Batch Size, 2]
You can use the read_bci_data function in the dataloader.py file to obtain the training data, training label, testing data and testing label.
train_data, train_label, test_data, test_label = read_bci_data()
In this project, Mean Squared Error and Crossentropy are the loss function, and Accuracy is the classification metrics.
- y_j: ground-truth value
- y_hat: predicted value from the regression model
- N: number of datums
- M: number of classes
- log: the natural log
- y: binary indicator (0 or 1) if class label c is the correct classification for observation o
- p: predicted probability observation o is of class c
- True Positive(TP) signifies how many positive class samples your model predicted correctly.
- True Negative(TN) signifies how many negative class samples your model predicted correctly.
- False Positive(FP) signifies how many negative class samples your model predicted incorrectly. This factor represents Type-I error in statistical nomenclature. This error positioning in the confusion matrix depends on the choice of the null hypothesis.
- False Negative(FN) signifies how many positive class samples your model predicted incorrectly. This factor represents Type-II error in statistical nomenclature. This error positioning in the confusion matrix also depends on the choice of the null hypothesis.
In the training step, there provided six file to train different model.
Each file contains a different model architecture with a different activation function. In addition, you can config the training parameters through the following argparse, and use the following instructions to train different method.
Finally, you will get such training result. The first picture is about DeepConvNet, and the second picture is about EEGNet.
You can get some detailed introduction and experimental results in this link.
parser.add_argument('--epochs', type=int, default='700', help='training epochs')
parser.add_argument('--learning_rate', type=float, default='1e-3', help='learning rate')
parser.add_argument('--save_model', action='store_true', help='check if you want to save the model.')
parser.add_argument('--save_csv', action='store_true', help='check if you want to save the training history.')
python DeepConvNet_training_ELU.py --epochs 3000 --learning_rate 1e-3 --save_model --save_csv
python DeepConvNet_training_LeakyReLU.py --epochs 3000 --learning_rate 1e-3 --save_model --save_csv
python DeepConvNet_training_ReLU.py --epochs 3000 --learning_rate 1e-3 --save_model --save_csv
python EEGNet_training_ELU.py --epochs 700 --learning_rate 1e-3 --save_model --save_csv
python EEGNet_training_LeakyReLU.py --epochs 700 --learning_rate 1e-3 --save_model --save_csv
python EEGNet_training_ReLU.py --epochs 700 --learning_rate 1e-3 --save_model --save_csv
You can display the testing results in different models by using the following commands in combination with different activation functions. The model checkpoint were in the checkpoint directory.
The detailed experimental result are in the link.
python model_testing.py
Then you will get the best result like this, each of the values were the testing accuracy.
| ReLU | LeakyReLU | ELU | |
|---|---|---|---|
| EEGNet | 87.1296 % | 88.2407 % | 87.2222 % |
| DeepConvNet | 85.4630 % | 84.0741 % | 83.7963 % |